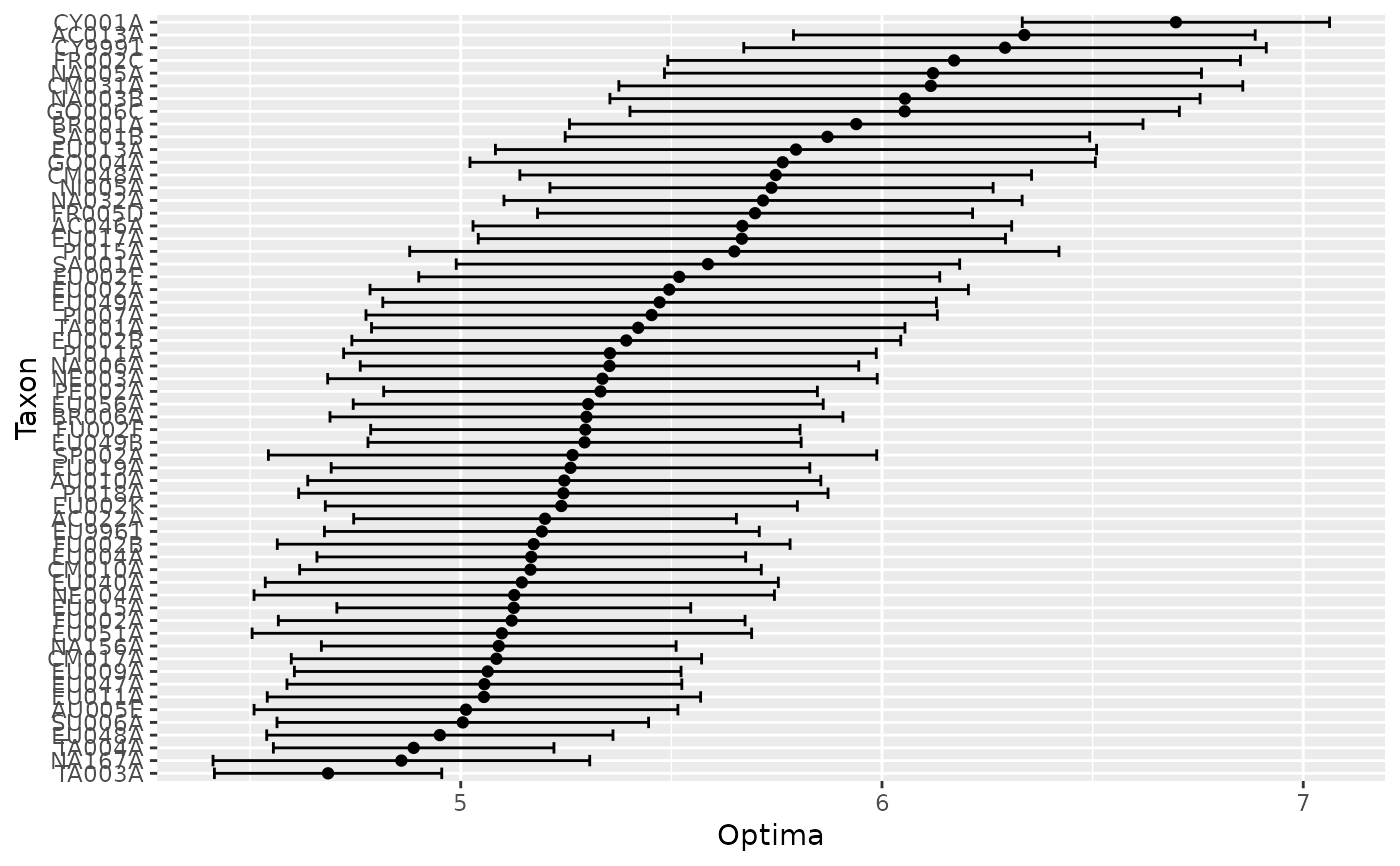
Plot of species WA optima and tolerance
Arguments
- x
A tolerance weighted weighted-average model from
WA- spp
data.frame of species data used to train the WA model
- minN2
numeric giving minimum N2 for inclusion in plot
- mult
numeric multiplier for the tolerances
Value
A ggplot object.
Details
Extracts and sorts WA optima and tolerances and
generates a ggplot.
Tends only to work well when there are a reasonable number of taxa,
otherwise it is difficult to read the names on the axis.
Rare taxa can be excluded with the minN2 argument.
The tol.cut argument in WA may need to be set to
prevent very small tolerances in rare taxa.
This function is very similar to the caterpillar
plot, but produces a ggplot
Examples
library(rioja)
data(SWAP)
mod <- WA(SWAP$spec, SWAP$pH, tolDW = TRUE)
coef(mod)
#> Optima Tolerances
#> AC001A 6.391870 0.70139514
#> AC002A 6.390380 0.53174428
#> AC004A 6.143205 0.55968148
#> AC013A 6.337889 0.54801922
#> AC014A 4.872948 0.49742581
#> AC014B 5.130048 0.37936515
#> AC014C 5.410085 0.45910379
#> AC017A 5.954913 0.82232953
#> AC018A 6.226881 0.78262599
#> AC019A 6.178677 0.48018379
#> AC022A 5.200095 0.45427325
#> AC025A 6.255318 0.63476241
#> AC028A 5.746221 0.56718098
#> AC029A 5.763416 0.67166841
#> AC030A 6.065965 0.56408377
#> AC034A 6.291338 0.48449928
#> AC035A 6.195437 0.62572120
#> AC039A 6.342105 0.69883268
#> AC042A 6.105013 0.56737545
#> AC044A 5.615805 0.65264916
#> AC046A 5.668511 0.63934402
#> AC048A 5.630603 0.47439600
#> AC9964 6.180631 0.63416440
#> AC9965 5.749658 0.38732095
#> AC9968 5.196932 0.28432793
#> AC9969 6.085539 0.54592485
#> AC9975 4.913026 0.30038533
#> AC9996 6.064632 0.44796754
#> AM001A 6.188654 0.88507076
#> AM001B 6.825107 0.25840115
#> AM001D 6.372088 0.52881939
#> AS001A 6.718995 0.39931168
#> AS003A 4.942885 0.45576886
#> AT001A 5.131810 0.32725815
#> AU001C 5.596840 0.72723945
#> AU002A 6.500123 0.49536843
#> AU003B 7.021573 0.93338074
#> AU003D 7.059654 0.41012188
#> AU004A 5.239846 0.74267814
#> AU004B 5.961804 0.75588138
#> AU004C 5.736427 0.90436855
#> AU004D 5.327831 0.44806407
#> AU005A 5.391870 0.64657405
#> AU005B 6.120225 0.92179682
#> AU005D 5.779317 0.67540958
#> AU005E 5.012466 0.50307312
#> AU005J 6.042784 0.63639617
#> AU005L 6.808868 0.26870066
#> AU009A 6.452710 0.40941482
#> AU009B 5.365533 0.37476674
#> AU010A 5.245831 0.60899533
#> AU010B 5.168809 0.44104019
#> AU014A 5.675902 0.54283987
#> AU022A 6.933716 1.06166819
#> AU023A 5.000000 0.55987045
#> AU9983 6.413181 0.60932418
#> AU9984 5.630266 0.57948634
#> AU9987 5.580902 0.47722157
#> AU9988 5.021488 0.28222996
#> BR001A 5.938892 0.68072286
#> BR003A 4.788705 0.37655266
#> BR004A 5.212293 0.56902740
#> BR005A 6.682530 0.42426400
#> BR006A 5.298437 0.60882266
#> BR9997 5.359548 0.60994450
#> CA018A 6.639938 0.07978854
#> CC001A 7.160000 0.55987045
#> CM004A 6.340316 0.67401573
#> CM010A 5.165520 0.54795242
#> CM013A 6.690273 0.37627094
#> CM014A 5.147159 0.29431575
#> CM015A 6.401941 0.58696795
#> CM015B 6.340000 0.18384792
#> CM017A 5.084739 0.48702456
#> CM020A 5.875921 0.58776954
#> CM031A 6.115911 0.74056374
#> CM031C 5.254492 0.36010767
#> CM038A 6.318335 1.11196218
#> CM048A 5.747689 0.60730927
#> CM050A 7.125273 0.34648216
#> CM051A 6.156664 0.62649361
#> CM052A 6.387611 0.66287223
#> CM101B 6.723777 0.73982106
#> CM9989 5.056829 0.50939149
#> CM9995 4.977483 0.37002637
#> CO001A 6.677402 0.32413464
#> CO001B 6.942000 0.55987045
#> CY001A 6.697727 0.36474275
#> CY002A 6.856985 0.17967760
#> CY003A 6.942000 0.55987045
#> CY004A 6.170910 0.56846466
#> CY007A 6.732602 0.41612477
#> CY010A 6.684801 0.34552697
#> CY013A 4.500000 0.55987045
#> CY9991 6.292045 0.62020591
#> DE001A 6.789421 0.41394315
#> DT002A 5.844772 0.41640861
#> DT003A 6.339250 0.44327754
#> DT004B 6.541467 0.55831524
#> EU002A 5.494897 0.71023015
#> EU002B 5.393005 0.65147688
#> EU002D 5.402762 0.43059249
#> EU002E 5.518692 0.61838477
#> EU002K 5.238856 0.56030659
#> EU003A 5.357044 0.78896659
#> EU004A 5.167489 0.50885666
#> EU009A 5.064030 0.45893425
#> EU009C 5.097029 0.41755953
#> EU011A 5.055075 0.51435907
#> EU013A 5.795851 0.71349014
#> EU014A 4.689540 0.29363389
#> EU015A 5.125939 0.42001561
#> EU016A 5.338856 0.55407170
#> EU017A 5.667389 0.62577660
#> EU019A 5.260537 0.56808478
#> EU020A 5.152957 0.50843332
#> EU021A 5.645646 0.50056773
#> EU022A 5.325298 0.51123442
#> EU025A 5.112205 0.85743165
#> EU027A 4.804029 0.41447365
#> EU028A 4.883259 0.38933030
#> EU028B 4.734235 0.29574483
#> EU031A 5.542663 0.67279398
#> EU034A 5.130405 0.46520539
#> EU039A 5.068189 0.58240247
#> EU040A 5.144991 0.60898198
#> EU046C 4.703086 0.26223116
#> EU047A 5.056132 0.46862621
#> EU048A 4.950387 0.41100932
#> EU049A 5.472081 0.65713660
#> EU049B 5.293949 0.51416261
#> EU051A 5.097642 0.59288269
#> EU051B 5.413136 0.51039524
#> EU056A 5.302624 0.55784186
#> EU057A 5.321476 0.48805222
#> EU058A 4.675294 0.19230803
#> EU9961 5.192557 0.51611301
#> EU9962 5.471678 0.61047017
#> EU9965 4.889374 0.36983461
#> EU9969 5.043360 0.15514991
#> FR001A 6.343205 0.58854774
#> FR001B 6.254040 1.22824447
#> FR002A 6.598967 0.63426291
#> FR002C 6.171183 0.67970724
#> FR005A 5.558934 0.87615633
#> FR005D 5.698662 0.51638499
#> FR006A 6.492085 0.47675595
#> FR007A 6.328192 0.62212664
#> FR008A 7.008896 0.23306195
#> FR009F 6.240000 0.55987045
#> FR010A 5.371060 0.45653330
#> FR011A 6.010973 0.51770904
#> FR015A 5.470155 0.60456010
#> FR018A 6.564130 0.53922033
#> FR9991 4.683372 0.32659560
#> FU002A 5.120969 0.55395297
#> FU002B 5.173162 0.60863688
#> FU002F 5.295851 0.50961554
#> GO003A 5.783958 0.58120235
#> GO004A 5.764200 0.74229530
#> GO006C 6.053800 0.65207382
#> GO013A 6.160208 0.64566976
#> GO023A 6.661725 0.55133685
#> GO025B 6.561954 0.55261231
#> GO025F 6.616286 1.18935372
#> GY005A 7.160000 0.55987045
#> HN001A 5.955994 0.66519957
#> ME019A 5.607095 0.67542207
#> NA002A 5.463491 0.75774912
#> NA003A 6.218315 0.77930741
#> NA003B 6.054642 0.70049398
#> NA005A 6.121030 0.63738270
#> NA005B 6.414172 0.60603115
#> NA006A 5.353125 0.59164936
#> NA006B 5.156900 0.60725281
#> NA007A 6.562142 0.43403153
#> NA008A 6.613639 0.41272741
#> NA013A 6.111603 0.57261545
#> NA014A 6.255649 0.64437758
#> NA015A 5.447810 0.72234339
#> NA016A 5.896107 0.73290874
#> NA032A 5.717606 0.61504150
#> NA033A 5.211043 0.64118526
#> NA037A 5.644886 0.59215006
#> NA038A 5.185541 0.60923474
#> NA042A 6.072514 0.52491794
#> NA043A 6.452144 0.46324591
#> NA044A 5.286432 0.45996184
#> NA045A 5.646395 0.57916186
#> NA046A 5.801026 0.70066300
#> NA048A 5.060848 0.37231488
#> NA063A 6.630172 0.20784694
#> NA068A 5.797265 0.70024649
#> NA084A 6.577000 0.55987045
#> NA086A 6.297220 0.54612594
#> NA099A 5.047937 0.55178862
#> NA101A 6.878816 0.90301989
#> NA102A 6.568537 0.71037023
#> NA112D 6.349672 0.33955919
#> NA113A 6.160973 0.77364704
#> NA114A 6.507217 0.41507194
#> NA115A 5.019151 0.39289907
#> NA129A 6.219000 0.59660650
#> NA133A 5.860350 0.68380996
#> NA135A 5.257653 0.25382779
#> NA140A 4.905698 0.38046078
#> NA149A 5.980457 0.60521923
#> NA151A 6.412941 0.83015405
#> NA156A 5.090226 0.42104814
#> NA158A 4.934526 0.29656636
#> NA160A 5.464112 0.53534889
#> NA167A 4.859099 0.44708153
#> NA170A 6.064604 0.57825328
#> NA9904 6.087969 0.57030330
#> NA9919 5.973559 0.77878714
#> NA9955 6.079392 0.66503459
#> NA9963 5.584184 0.53528847
#> NA9964 5.319150 0.59563314
#> NE003A 5.336385 0.65225852
#> NE003B 5.017068 0.45396423
#> NE003C 5.651123 0.56114888
#> NE004A 5.127014 0.61767941
#> NE012A 5.484129 0.41560705
#> NE020A 5.451155 0.47949394
#> NE023A 5.738833 0.77171903
#> NI002A 6.341801 0.65310672
#> NI005A 5.737781 0.52599353
#> NI008A 6.395290 0.38001995
#> NI009A 6.239170 0.59620530
#> NI009B 6.169422 0.49942808
#> NI017A 5.877377 0.50445434
#> NI021A 5.949433 0.64259959
#> NI026A 6.295821 0.67061947
#> NI027A 6.546756 0.33566555
#> NI152A 5.813076 0.65260496
#> NI9984 7.250000 0.55987045
#> OP001A 6.465227 0.56595221
#> PE002A 5.331847 0.51478204
#> PI005A 5.269995 0.58474691
#> PI007A 5.453201 0.67814977
#> PI011A 5.354191 0.63224739
#> PI014A 5.833683 0.68715726
#> PI015A 5.649451 0.77063763
#> PI016A 5.929813 0.68462607
#> PI018A 5.243574 0.62838266
#> PI018B 5.331454 0.54572164
#> PI022B 4.963960 0.48426796
#> PI023A 5.350816 0.72884892
#> PI055A 6.600000 0.55987045
#> PI056A 4.895904 0.55460964
#> PI139A 5.517000 0.55987045
#> PI164A 5.505341 0.75028058
#> RH006B 6.258497 1.19359605
#> SA001A 5.586822 0.59756141
#> SA001B 5.870518 0.62257278
#> SA006A 5.924093 0.84900971
#> SA042A 5.056502 0.56501329
#> SE001A 4.764743 0.35429597
#> SP002A 5.265415 0.72201427
#> ST004A 6.860809 0.23750553
#> ST010A 7.093000 0.55987045
#> SU002A 5.517000 0.55987045
#> SU004A 5.321936 0.45528067
#> SU005A 5.322800 0.66478302
#> SU006A 5.004872 0.44104569
#> SY002A 6.435810 0.60275162
#> SY003A 6.430885 0.49001569
#> SY004A 6.687533 0.46853936
#> SY009A 6.248336 0.88788215
#> SY010A 6.044152 0.66824846
#> SY013A 6.587861 0.69863514
#> SY043A 5.894069 0.55295755
#> TA001A 5.421338 0.63313074
#> TA002A 5.865716 0.74840640
#> TA003A 4.685068 0.26982689
#> TA004A 4.888238 0.33300474
#> TA9996 6.069182 0.76064536
centipede_plot(mod, spp = SWAP$spec, minN2 = 20)